Co-Project leader: Dr. Mary-Ann Blätke (IPK)
Email:WP II.8 (Metabolic Networks) aims to develop a testable metabolic network of B. napus, including essential aspects of lipid metabolism. The metabolic network approach relies on stoichiometric modelling (also known as flux balance analyses). This approach requires a precise metabolic map of rapeseed seed metabolism, input fluxes, and output fluxes, as well as a set of constraints with approximate seed conditions. Depending on the optimisation function, it may resolve how metabolism proceeds most efficiently with respect to energy and/or nitrogen investment and/or output optimisation.
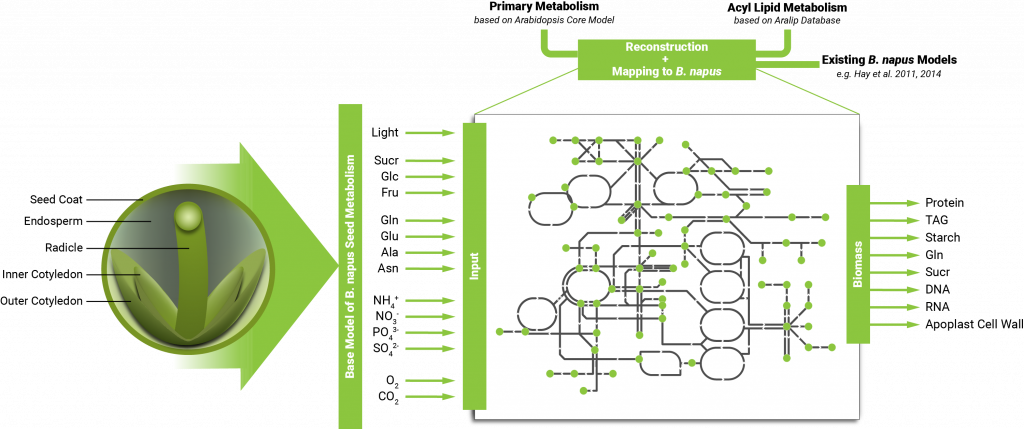
We will generate and integrate a rapeseed specific bottom-up metabolic maps of primary carbon and nitrogen metabolism and detailed lipid metabolism in a first step. For all reactions where information is available in any plant species, the likely flux carrying enzyme(s) or transport protein(s) will be identified in the B. napus and Arabidopsis thaliana genome for comparison to earlier publications. Inputs will be calculated from leaf petiole output to approximate a typical dicot plant phloem content. Outputs will be coded as seed storage contents in oil, protein and starch for temporary storage. The data generated in the project will be used to (i) constrain the models and test for changes in output vs the idealised solution, to (ii) identify the reactions which control flux, and to (iii) visualise fluxes in space and time using 3D technology.
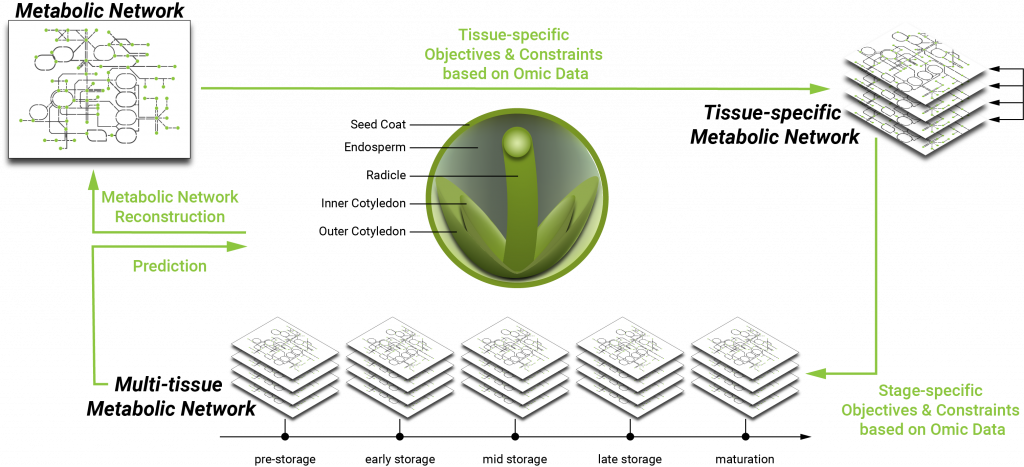
The in silico experiments based on the integrated rapeseed data strive to analyse the impact of resource nutrients, environmental factors, the expression of genetic information (transcriptome, proteome) on seed development, and thus on the phenotypic traits that highly depend on the state of metabolism. The analysis will help to elucidate the genome-phenome relationship by correlating observed metabolic flux distributions to phenotypic traits.